MoveGenerator contains parent classes for all move generators. A MoveGenerator sub-class is used at fullrmc’s stochastic engine runtime to generate moves upon selected groups. Every group has its own MoveGenerator class and definitions, therefore it is possible to fully customize how a group of atoms should move.
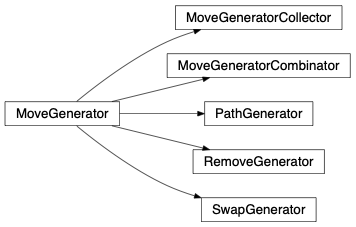
fullrmc.Core.MoveGenerator.MoveGenerator(group=None)Bases: object
It is the parent class of all moves generators. This class can’t be instantiated but its sub-classes might be.
| Parameters: |
|
|---|
groupGroup instance.
set_group(group)Set the MoveGenerator group.
| Parameters: |
|
|---|
check_group(group)Check the generator’s group. This method must be overloaded in all MoveGenerator sub-classes.
| Parameters: |
|
|---|
transform_coordinates(coordinates, argument=None)Transform coordinates. This method is called to move atoms. This method must be overloaded in all MoveGenerator sub-classes.
| Parameters: |
|
|---|---|
| Returns: |
|
move(coordinates)Moves coordinates. This method must NOT be overloaded in MoveGenerator sub-classes.
| Parameters: |
|
|---|---|
| Returns: |
|
fullrmc.Core.MoveGenerator.RemoveGenerator(group=None, maximumCollected=None, allowFittingScaleFactor=False, atomsList=None)Bases: fullrmc.Core.MoveGenerator.MoveGenerator
This is a very particular move generator that will not generate moves on atoms but removes them from the atomic configuration using a general collector mechanism. Remove generators must be used to create defects in the simulated system. When the standard error is high, removing atoms might reduce the total fit standard error but this can be illusional and very limiting because artificial non physical voids can get created in the system which will lead to an impossibility to finding a solution at the end. It’s strongly recommended to exhaust all ideas and possibilities in finding a good solution prior to start removing atoms unless structural defects is the goal of the simulation.
All removed or amputated atoms are collected by the engine and will become available to be re-inserted in the system if needed. But keep in mind, it might be physically easy to remove and atom but an impossibility to add it back especially if the created voids are smeared out.
Removers are called generators but they behave like selectors. Instead of applying a certain move on a group of atoms, they normally pick atoms from defined atoms list and apply no moves on those. ‘move’ and ‘transform_coordinates’ methods are not implemented in this class of generators and a usage error will be raised if called. ‘pick_from_list’ method is used instead and must be overloaded by all RemoveGenerator subclasses.
N.B. This class can’t be instantiated but its sub-classes might be.
| Parameters: |
|
|---|
atomsListAtoms list from which atoms will be picked to attempt removal.
allowFittingScaleFactorWhether to allow constraints to fit their scale factor upon removing atoms.
maximumCollectedMaximum collected atoms allowed.
check_group(group)Check the generator’s group.
| Parameters: |
|
|---|
set_maximum_collected(maximumCollected)Set maximum collected number of atoms allowed.
| Parameters: |
|
|---|
set_allow_fitting_scale_factor(allowFittingScaleFactor)Set allow fitting scale factor flag.
| Parameters: |
|
|---|
set_atoms_list(atomsList)Set atoms index list from which atoms will be picked to attempt removal. This method must be overloaded and not be called from this class but from its children. Otherwise a usage error will be raised.
| Parameters: |
|
|---|
move(coordinates)Moves coordinates. This method must NOT be overloaded in MoveGenerator sub-classes.
| Parameters: |
|
|---|
transform_coordinates(coordinates, argument)This method must NOT be overloaded in MoveGenerator sub-classes.
| Parameters: |
|
|---|
pick_from_list(engine)This method must be overloaded in all RemoveGenerator sub-classes.
| Parameters: |
|
|---|
fullrmc.Core.MoveGenerator.SwapGenerator(group=None, swapLength=1, swapList=None)Bases: fullrmc.Core.MoveGenerator.MoveGenerator
It is a particular move generator that instead of generating a move upon a group of atoms, it will exchange the group atom positions with other atoms from a defined swapList. Because the swapList can be big, swapGenerator can be assigned to multiple groups at the same time under the condition of all groups having the same length.
During stochastic engine runtime, whenever a swap generator is encountered, all sophisticated selection recurrence modes such as (refining, exploring) will be reduced to simple recurrence.
This class can’t be instantiated but its sub-classes might be.
| Parameters: |
|
|---|
swapLengthSwap length.
swapListSwap list.
groupAtomsIndexesLast selected group atoms index.
swapAtomsIndexesLast swap atoms index.
set_swap_length(swapLength)Set swap length. it will empty and reset swaplist automatically.
| Parameters: |
|
|---|
set_group(group)Set the MoveGenerator group.
| Parameters: |
|
|---|
set_swap_list(swapList)Set the swap-list to exchange atoms position from.
| Parameters: |
|
|---|
append_to_swap_list(subList)Append a sub list to swap list.
| Parameters: |
|
|---|
get_ready_for_move(engine, groupAtomsIndexes)Set the swap generator ready to perform a move. Unlike a normal move generator, swap generators will affect not only the selected atoms but other atoms as well. Therefore at stochastic engine runtime, selected atoms will be extended to all affected atoms by the swap.
This method is called automatically upon stochastic engine runtime to ensure that all affect atoms with the swap are updated.
| Parameters: |
|
|---|---|
| Returns: |
|
fullrmc.Core.MoveGenerator.PathGenerator(group=None, path=None, randomize=False)Bases: fullrmc.Core.MoveGenerator.MoveGenerator
PathGenerator is a MoveGenerator sub-class where moves definitions are pre-stored in a path and get pulled out at every move step.
This class can’t be instantiated but its sub-classes might be.
| Parameters: |
|
|---|
stepCurrent step number.
pathPath list of moves.
randomizeRandomize flag.
check_path(path)Check the generator’s path.
This method must be overloaded in all PathGenerator sub-classes.
| Parameters: |
|
|---|
normalize_path(path)Normalizes all path moves. It is called automatically upon set_path method is called.
This method can be overloaded in all MoveGenerator sub-classes.
| Parameters: |
|
|---|---|
| Returns: |
|
set_path(path)Set the moves path.
| Parameters: |
|
|---|
set_randomize(randomize)Set whether to randomize moves selection.
| Parameters: |
|
|---|
move(coordinates)Move coordinates.
| Parameters: |
|
|---|---|
| Returns: |
|
fullrmc.Core.MoveGenerator.MoveGeneratorCombinator(group=None, combination=None, shuffle=False)Bases: fullrmc.Core.MoveGenerator.MoveGenerator
MoveGeneratorCombinator combines all moves of a list of MoveGenerators and applies it at once.
| Parameters: |
|
|---|
# import fullrmc modules
from fullrmc.Engine import Engine
from fullrmc.Core.MoveGenerator import MoveGeneratorCombinator
from fullrmc.Generators.Translations import TranslationGenerator
from fullrmc.Generators.Rotations import RotationGenerator
# create engine
ENGINE = Engine(path='my_engine.rmc')
# set pdb file
ENGINE.set_pdb('system.pdb')
# Add constraints ...
# Re-define groups if needed ...
##### Define each group move generator as a combination of a translation and a rotation. #####
# create recursive group selector. Recurrence is set to 20 with explore flag set to True.
# shuffle is set to True which means that at every selection the order of move generation
# is random. At one step a translation is performed prior to rotation and in another step
# the rotation is performed at first.
# selected from the collector.
for g in ENGINE.groups:
# create translation generator
TMG = TranslationGenerator(amplitude=0.2)
# create rotation generator only when group length is bigger than 1.
if len(g)>1:
RMG = RotationGenerator(amplitude=2)
MG = MoveGeneratorCombinator(collection=[TMG,RMG],shuffle=True)
else:
MG = MoveGeneratorCombinator(collection=[TMG],shuffle=True)
g.set_move_generator( MG )
shuffleShuffle flag.
combinationCombination list of MoveGenerator instances.
check_group(group)Checks the generator’s group. This methods always returns True because normally all combination MoveGenerator instances groups are checked.
This method must NOT be overloaded unless needed.
| Parameters: |
|
|---|
set_group(group)Set the MoveGenerator group.
| Parameters: |
|
|---|
set_combination(combination)Set the generators combination list.
| Parameters: |
|
|---|
set_shuffle(shuffle)Set whether to shuffle moves generator.
| Parameters: |
|
|---|
move(coordinates)Move coordinates.
| Parameters: |
|
|---|---|
| Returns: |
|
fullrmc.Core.MoveGenerator.MoveGeneratorCollector(group=None, collection=None, randomize=True, weights=None)Bases: fullrmc.Core.MoveGenerator.MoveGenerator
MoveGeneratorCollector collects MoveGenerators instances and applies the move of one instance at every step.
| Parameters: |
|
|---|
# import fullrmc modules
from fullrmc.Engine import Engine
from fullrmc.Core.MoveGenerator import MoveGeneratorCollector
from fullrmc.Generators.Translations import TranslationGenerator
from fullrmc.Generators.Rotations import RotationGenerator
# create engine
ENGINE = Engine(path='my_engine.rmc')
# set pdb file
ENGINE.set_pdb('system.pdb')
# Add constraints ...
# Re-define groups if needed ...
##### Define each group move generator as a combination of a translation and a rotation. #####
# create recursive group selector. Recurrence is set to 20 with explore flag set to True.
# randomize is set to True which means that at every selection a generator is randomly
# selected from the collector.
for g in ENGINE.groups:
# create translation generator
TMG = TranslationGenerator(amplitude=0.2)
# create rotation generator only when group length is bigger than 1.
if len(g)>1:
RMG = RotationGenerator(amplitude=2)
MG = MoveGeneratorCollector(collection=[TMG,RMG],randomize=True)
else:
MG = MoveGeneratorCollector(collection=[TMG],randomize=True)
g.set_move_generator( MG )
randomizeRandomize flag.
collectionList of MoveGenerator instances.
generatorsWeightGenerators selection weights list.
selectionSchemeSelection scheme.
set_group(group)Set the MoveGenerator group.
| Parameters: |
|
|---|
check_group(group)Check the generator’s group. This methods always returns True because normally all collection MoveGenerator instances groups are checked.
This method must NOT be overloaded unless needed.
| Parameters: |
|
|---|
set_collection(collection)Set the generators instances collection list.
| Parameters: |
|
|---|
set_randomize(randomize)Set whether to randomize MoveGenerator instance selection from collection list.
| Parameters: |
|
|---|
set_weights(weights)Set groups selection weighting scheme.
| Parameters: |
|
|---|
set_selection_scheme()Set selection scheme.
move(coordinates)Move coordinates.
| Parameters: |
|
|---|---|
| Returns: |
|
fullrmc.Core.MoveGenerator.generate_random_float()random() -> x in the interval [0, 1).
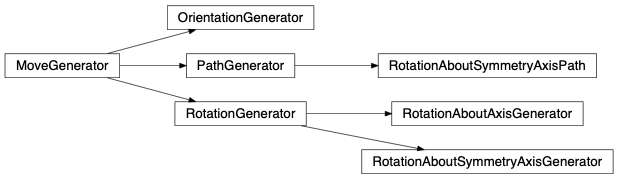
Rotations contains all rotation like MoveGenerator classes.
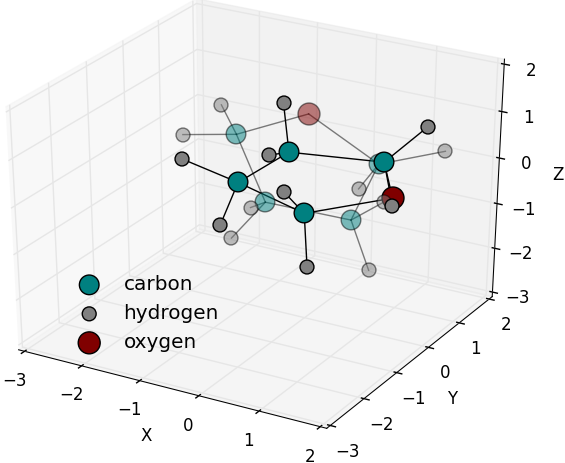
Random rotation axis and angle generated and
applied on a Tetrahydrofuran molecule. Solid
colours are of the origin molecule position while
fading ones are of the rotated molecule.
( |

Random rotation generated about a pre-defined axis
or one of the symmetry axes of the Tetrahydrofuran
molecule. Solid colours are of the origin molecule
position while fading ones are of the rotated
molecule.
( |
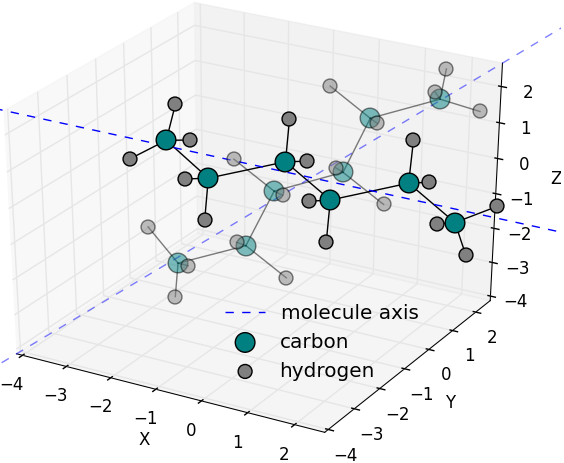
Random orientation of hexane molecule to [1,1,1]
axis with maximumOffsetAngle of 10 degrees is
generated. First principal axis of hexane molecule
is considered as groupAxis. Solid colors are of
original molecule while fading ones are of the
oriented one. ( |
fullrmc.Generators.Rotations.RotationGenerator(group=None, amplitude=2)¶Bases: fullrmc.Core.MoveGenerator.MoveGenerator
Generate random rotational moves upon groups of atoms. Only groups of more than one atom are accepted.
| Parameters: |
|
|---|
# import fullrmc modules
from fullrmc.Engine import Engine
from fullrmc.Generators.Rotations import RotationGenerator
# create engine
ENGINE = Engine(path='my_engine.rmc')
# set pdb file
ENGINE.set_pdb('system.pdb')
# Add constraints ...
# Re-define groups if needed ...
# Re-define groups selector if needed ...
# set moves generators to random rotations.
# Maximum rotation amplitude is set to 5 degrees to all defined groups
for g in ENGINE.groups:
if len(g) >1:
g.set_move_generator( RotationGenerator(amplitude=5) )
amplitude¶Maximum allowed angle of rotation in rad.
set_amplitude(amplitude)¶Set maximum rotation angle in degrees and transforms it to rad.
| Parameters: |
|
|---|
check_group(group)¶Check the generator’s group.
| Parameters: |
|
|---|
transform_coordinates(coordinates, argument=None)¶Rotate coordinates.
| Parameters: |
|
|---|---|
| Returns: |
|
fullrmc.Generators.Rotations.RotationAboutAxisGenerator(group=None, amplitude=2, axis=(1, 0, 0))¶Bases: fullrmc.Generators.Rotations.RotationGenerator
Generates random rotational moves upon groups of atoms about a pre-defined axis.
| Parameters: |
|
|---|
# import fullrmc modules
from fullrmc.Engine import Engine
from fullrmc.Generators.Rotations import RotationAboutAxisGenerator
# create engine
ENGINE = Engine(path='my_engine.rmc')
# set pdb file
ENGINE.set_pdb('system.pdb')
# Add constraints ...
# Re-define groups if needed ...
# Re-define groups selector if needed ...
# set moves generators to random rotations about (1,1,1) a pre-defined axis.
# Maximum rotation amplitude is set to 5 degrees to all defined groups
for g in ENGINE.groups:
if len(g) >1:
g.set_move_generator( RotationAboutAxisGenerator(amplitude=5, axis=(1,1,1)) )
axis¶Rotation axis vector.
check_group(group)¶Check the generator’s group.
| Parameters: |
|
|---|
set_axis(axis)¶Set the axis along which the rotation will be performed.
| Parameters: |
|
|---|
transform_coordinates(coordinates, argument=None)¶Rotate coordinates.
| Parameters: |
|
|---|---|
| Returns: |
|
fullrmc.Generators.Rotations.RotationAboutSymmetryAxisGenerator(group=None, amplitude=2, axis=0)¶Bases: fullrmc.Generators.Rotations.RotationGenerator
Generates random rotational moves upon groups of atoms about one of their symmetry axis. Only groups of more than 1 atom are accepted.
| Parameters: |
|
|---|
# import fullrmc modules
from fullrmc.Engine import Engine
from fullrmc.Generators.Rotations import RotationAboutSymmetryAxisGenerator
# create engine
ENGINE = Engine(path='my_engine.rmc')
# set pdb file
ENGINE.set_pdb('system.pdb')
# Add constraints ...
# Re-define groups if needed ...
# Re-define groups selector if needed ...
# set moves generators to random rotations about the second symmetry axis of each group.
# Maximum rotation amplitude is set to 5 degrees to all defined groups
for g in ENGINE.groups:
if len(g) >1:
g.set_move_generator( RotationAboutSymmetryAxisGenerator(amplitude=5, axis=1) )
axis¶Rotation axis index.
set_axis(axis)¶Set the symmetry axis index to rotate about.
| Parameters: |
|
|---|
transform_coordinates(coordinates, argument=None)¶Rotate coordinates.
| Parameters: |
|
|---|---|
| Returns: |
|
fullrmc.Generators.Rotations.RotationAboutSymmetryAxisPath(group=None, axis=0, path=None, randomize=False)¶Bases: fullrmc.Core.MoveGenerator.PathGenerator
Generate rotational moves upon groups of atoms about one of their symmetry axis. Only groups of more than one atom are accepted.
| Parameters: |
|
|---|
# import fullrmc modules
from fullrmc.Engine import Engine
from fullrmc.Generators.Rotations import RotationAboutSymmetryAxisPath
# create engine
ENGINE = Engine(path='my_engine.rmc')
# set pdb file
ENGINE.set_pdb('system.pdb')
# Add constraints ...
# Re-define groups if needed ...
# Re-define groups selector if needed ...
# set moves generators to pre-defined rotations about the second symmetry axis of each group.
angles = [-0.1, -0.5, -0.05, 0.5, 0.01, 2, 3, 1, -3]
for g in ENGINE.groups:
if len(g) >1:
g.set_move_generator( RotationAboutSymmetryAxisPath(axis=1, path=angles) )
axis¶Rotation axis index.
set_axis(axis)¶Set the symmetry axis index to rotate about.
| Parameters: |
|
|---|
check_path(path)¶Check the generator’s path.
| Parameters: |
|
|---|
normalize_path(path)¶Transform all path angles to radian.
| Parameters: |
|
|---|---|
| Returns: |
|
check_group(group)¶Check the generator’s group.
| Parameters: |
|
|---|
transform_coordinates(coordinates, argument)¶Rotate coordinates.
| Parameters: |
|
|---|---|
| Returns: |
|
fullrmc.Generators.Rotations.OrientationGenerator(group=None, maximumOffsetAngle=10, groupAxis={'symmetry': 0}, orientationAxis={'fixed': (1, 0, 0)}, flip=None)¶Bases: fullrmc.Core.MoveGenerator.MoveGenerator
Generate rotational moves upon groups of atoms to align and orient along an axis. Orientation rotations are computed randomly allowing offset angle between grouAxis and orientationAxis Only groups of more than 1 atom are accepted.
| Parameters: |
|
|---|
# import fullrmc modules
from fullrmc.Engine import Engine
from fullrmc.Generators.Rotations import OrientationGenerator
# create engine
ENGINE = Engine(path='my_engine.rmc')
# set pdb file
ENGINE.set_pdb('system.pdb')
# Add constraints ...
# Re-define groups if needed ...
# Re-define groups selector if needed ...
# set moves generators to orientations of each group third symmetry axis
# towards the (-1,0,2) the predefined axis within maximum 5 degrees.
for g in ENGINE.groups:
if len(g) >1:
g.set_move_generator( OrientationGenerator(maximumOffsetAngle=5,
groupAxis={"symmetry":2},
orientationAxis={"fixed":(-1,0,2)}) )
maximumOffsetAngle¶Maximum offset angle in degrees between groupAxis and orientationAxis in rad.
orientationAxis¶Orientation axis value or definition.
groupAxis¶Group axis value or definition.
flip¶Flip value.
set_maximum_offset_angle(maximumOffsetAngle)¶Set the maximum offset angle allowed.
| Parameters: |
|
|---|
check_group(group)¶Check the generator’s group.
| Parameters: |
|
|---|
set_flip(flip)¶Set flip flag value.
| Parameters: |
|
|---|
set_group_axis(groupAxis)¶Sets group axis value.
| Parameters: |
|
|---|
set_orientation_axis(orientationAxis)¶Set orientation axis value.
| Parameters: |
|
|---|
transform_coordinates(coordinates, argument=None)¶Rotate coordinates.
| Parameters: |
|
|---|---|
| Returns: |
|
fullrmc.Generators.Rotations.generate_random_float()¶random() -> x in the interval [0, 1).
Translations contains all translation like MoveGenerator classes.
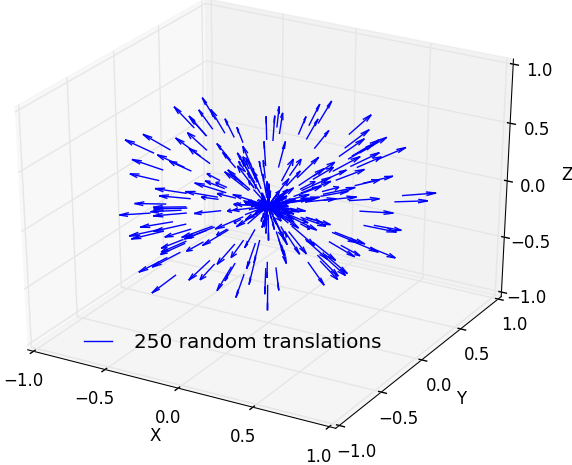
Random translation vectors generated from
atom at origin. ( |
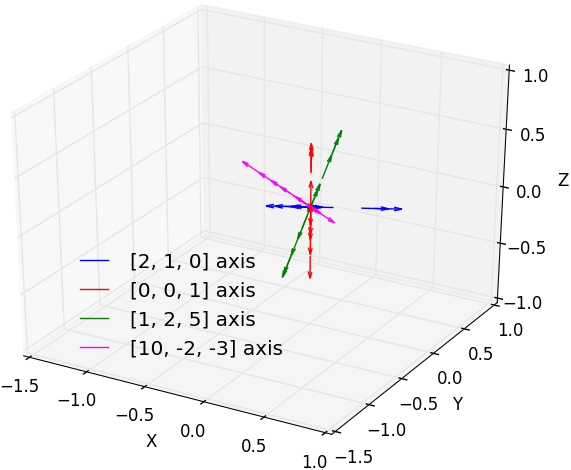
Random translation vectors generated from
atom at origin along a pre-defined axis.
( |
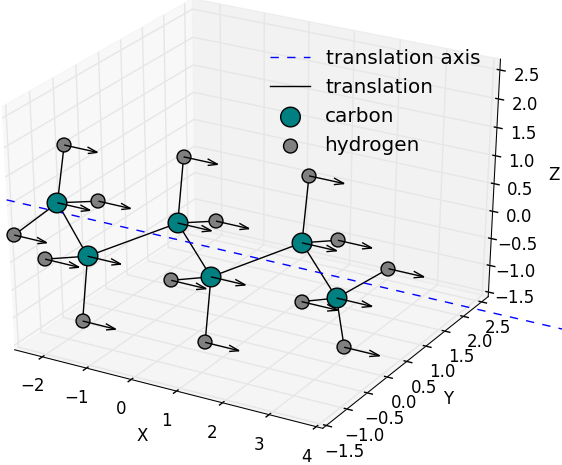
Random translation vector generated along a
predefined axis or one of the symmetry axes of the
hexane molecule and applied on all the molecule’s
atoms at the same time.
( |
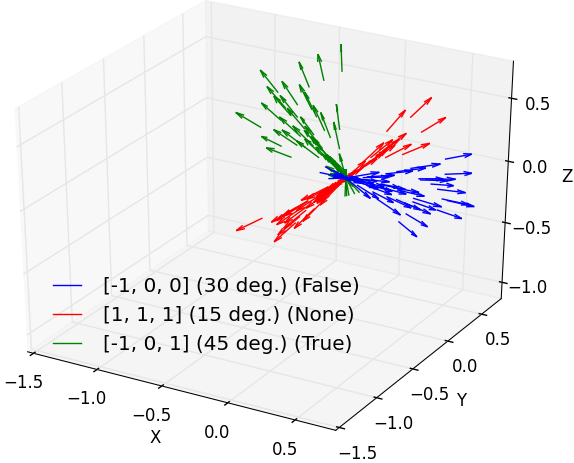
Random translation vectors generated towards an
axis within some maximum angle.
Legend is formatted as axis (angle) (direction)
( |
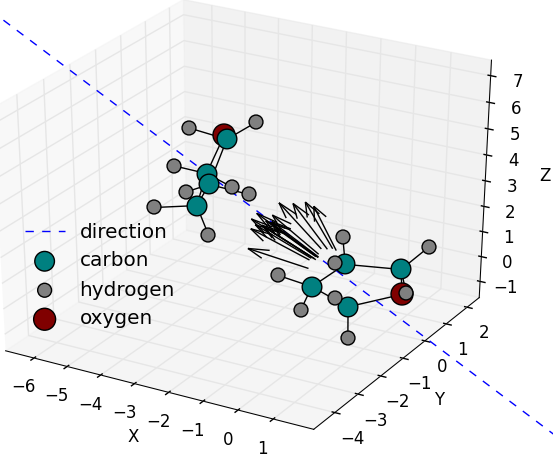
Random translation vectors generated towards a
pre-defined center or towards the geometric center
of a group of atoms. Here 20 vectors are generated
within a maximum separation angle of 30 deg.
( |
fullrmc.Generators.Translations.TranslationGenerator(group=None, amplitude=0.2)¶Bases: fullrmc.Core.MoveGenerator.MoveGenerator
Generates random translations moves upon groups of atoms.
| Parameters: |
|
|---|
# import fullrmc modules
from fullrmc.Engine import Engine
from fullrmc.Generators.Translations import TranslationGenerator
# create engine
ENGINE = Engine(path='my_engine.rmc')
# set pdb file
ENGINE.set_pdb('system.pdb')
# Add constraints ...
# Re-define groups if needed ...
# Re-define groups selector if needed ...
# set moves generators to random translations.
# Maximum translation amplitude is set to 0.3A to all defined groups
for g in ENGINE.groups:
g.set_move_generator( TranslationGenerator(amplitude=0.3) )
amplitude¶Translation amplitude limits.
set_amplitude(amplitude)¶Set maximum translation vector allowed amplitude.
| Parameters: |
|
|---|
check_group(group)¶Check the generator’s group.
| Parameters: |
|
|---|
transform_coordinates(coordinates, argument=None)¶Translate coordinates.
| Parameters: |
|
|---|---|
| Returns: |
|
fullrmc.Generators.Translations.TranslationAlongAxisGenerator(group=None, amplitude=0.2, axis=(1, 0, 0), direction=None)¶Bases: fullrmc.Generators.Translations.TranslationGenerator
Generates random translation moves upon groups of atoms along a pre-defined axis.
| Parameters: |
|
|---|
# import fullrmc modules
from fullrmc.Engine import Engine
from fullrmc.Generators.Translations import TranslationAlongAxisGenerator
# create engine
ENGINE = Engine(path='my_engine.rmc')
# set pdb file
ENGINE.set_pdb('system.pdb')
# Add constraints ...
# Re-define groups if needed ...
# Re-define groups selector if needed ...
# set moves generators to translations along pre-defined axis (1,1,1).
# Maximum translation amplitude is set to 0.3A to all defined groups
for g in ENGINE.groups:
g.set_move_generator( TranslationAlongAxisGenerator(amplitude=0.3, axis=(1,1,1)) )
axis¶Translation axis.
direction¶Generated translation vectors direction.
set_axis(axis)¶Set the axis along which the translation will be performed.
| Parameters: |
|
|---|
set_direction(direction)¶Sets the generated translation vectors direction.
| Parameters: |
|
|---|
transform_coordinates(coordinates, argument=None)¶translates coordinates.
| Parameters: |
|
|---|---|
| Returns: |
|
fullrmc.Generators.Translations.TranslationTowardsAxisGenerator(group=None, amplitude=0.2, axis=(1, 0, 0), angle=30, direction=True)¶Bases: fullrmc.Generators.Translations.TranslationAlongAxisGenerator
Generates random translation moves upon groups of atoms towards a pre-defined axis within a tolerance angle between translation vectors and the pre-defined axis.
| Parameters: |
|
|---|
# import fullrmc modules
from fullrmc.Engine import Engine
from fullrmc.Generators.Translations import TranslationTowardsAxisGenerator
# create engine
ENGINE = Engine(path='my_engine.rmc')
# set pdb file
ENGINE.set_pdb('system.pdb')
# Add constraints ...
# Re-define groups if needed ...
# Re-define groups selector if needed ...
# set moves generators to translations towards a pre-defined axis (1,1,1) within 10 degrees.
# Maximum translation amplitude is set to 0.3A to all defined groups
for g in ENGINE.groups:
g.set_move_generator( TranslationTowardsAxisGenerator(amplitude=0.3, axis=(1,1,1), angle=10) )
angle¶Tolerance maximum angle in rad.
set_angle(angle)¶Set the tolerance maximum angle.
| Parameters: |
|
|---|
transform_coordinates(coordinates, argument=None)¶translate coordinates.
| Parameters: |
|
|---|---|
| Returns: |
|
fullrmc.Generators.Translations.TranslationAlongSymmetryAxisGenerator(group=None, amplitude=0.2, axis=0, direction=None)¶Bases: fullrmc.Generators.Translations.TranslationGenerator
Generate random translation moves upon groups of atoms along one of their symmetry axis. Only groups containing more than 1 atoms allow computing symmetry axis.
| Parameters: |
|
|---|
# import fullrmc modules
from fullrmc.Engine import Engine
from fullrmc.Generators.Translations import TranslationAlongSymmetryAxisGenerator
# create engine
ENGINE = Engine(path='my_engine.rmc')
# set pdb file
ENGINE.set_pdb('system.pdb')
# Add constraints ...
# Re-define groups if needed ...
# Re-define groups selector if needed ...
# set moves generators to translations along the second symmetry axis of every group.
# Maximum translation amplitude is set to 0.3A to all defined groups.
for g in ENGINE.groups:
if len(g)>1:
g.set_move_generator( TranslationAlongSymmetryAxisGenerator(amplitude=0.3, axis=(1,1,1), axis=1) )
axis¶Translation axis index.
direction¶Generated translation vectors direction.
check_group(group)¶Check the generator’s group.
| Parameters: |
|
|---|
set_axis(axis)¶Set the symmetry axis index to translate along.
| Parameters: |
|
|---|
set_direction(direction)¶Sets the generated translation vectors direction.
| Parameters: |
|
|---|
transform_coordinates(coordinates, argument=None)¶translate coordinates.
| Parameters: |
|
|---|---|
| Returns: |
|
fullrmc.Generators.Translations.TranslationTowardsSymmetryAxisGenerator(group=None, amplitude=0.2, axis=0, angle=30, direction=True)¶Bases: fullrmc.Generators.Translations.TranslationAlongSymmetryAxisGenerator
Generates random translation moves upon groups of atoms towards one of its symmetry axis within a tolerance angle between translation vectors and the axis. Only groups of more than 1 atom are accepted.
| Parameters: |
|
|---|
# import fullrmc modules
from fullrmc.Engine import Engine
from fullrmc.Generators.Translations import TranslationTowardsSymmetryAxisGenerator
# create engine
ENGINE = Engine(path='my_engine.rmc')
# set pdb file
ENGINE.set_pdb('system.pdb')
# Add constraints ...
# Re-define groups if needed ...
# Re-define groups selector if needed ...
# set moves generators to translations towards the first symmetry axis of every group within 15 degrees.
# Maximum translation amplitude is set to 0.3A to all defined groups.
for g in ENGINE.groups:
if len(g)>1:
g.set_move_generator( TranslationTowardsSymmetryAxisGenerator(amplitude=0.3, axis=0, angle=15) )
angle¶Tolerance maximum angle in rad.
set_angle(angle)¶Set the tolerance maximum angle.
| Parameters: |
|
|---|
transform_coordinates(coordinates, argument=None)¶translate coordinates.
| Parameters: |
|
|---|---|
| Returns: |
|
fullrmc.Generators.Translations.TranslationAlongSymmetryAxisPath(group=None, axis=0, path=None, randomize=False)¶Bases: fullrmc.Core.MoveGenerator.PathGenerator
Generates translation moves upon groups of atoms along one of their symmetry axis. Only groups of more than 1 atom are accepted.
| Parameters: |
|
|---|
# import fullrmc modules
from fullrmc.Engine import Engine
from fullrmc.Generators.Translations import TranslationAlongSymmetryAxisPath
# create engine
ENGINE = Engine(path='my_engine.rmc')
# set pdb file
ENGINE.set_pdb('system.pdb')
# Add constraints ...
# Re-define groups if needed ...
# Re-define groups selector if needed ...
# set moves generators to translations of predefined amplitudes along the first symmetry axis of every group.
amps = [-0.1, 0.075, -0.05, -0.25, 0.01, 0.02, 0.03, 0.1, 0.3]
for g in ENGINE.groups:
if len(g)>1:
g.set_move_generator( TranslationAlongSymmetryAxisPath(axis=0, path=amps) )
axis¶Translation axis index.
check_group(group)¶Check the generator’s group.
| Parameters: |
|
|---|
set_axis(axis)¶Set the symmetry axis index to translate along.
| Parameters: |
|
|---|
check_path(path)¶Check the generator’s path.
| Parameters: |
|
|---|
normalize_path(path)¶Transforms all path distances to floating numbers.
| Parameters: |
|
|---|---|
| Returns: |
|
transform_coordinates(coordinates, argument)¶Translate coordinates.
| Parameters: |
|
|---|---|
| Returns: |
|
fullrmc.Generators.Translations.TranslationTowardsCenterGenerator(group=None, center={'fixed': (0, 0, 0)}, amplitude=0.1, angle=30, direction=True)¶Bases: fullrmc.Generators.Translations.TranslationGenerator
Generates random translation moves of every atom of the group along its direction vector to the geometric center of the group.
| Parameters: |
|
|---|
# import fullrmc modules
from fullrmc.Engine import Engine
from fullrmc.Generators.Translations import TranslationTowardsCenterGenerator
# create engine
ENGINE = Engine(path='my_engine.rmc')
# set pdb file
ENGINE.set_pdb('system.pdb')
# Add constraints ...
# Re-define groups if needed ...
# Re-define groups selector if needed ...
# set moves generators to translations towards the origin defined as (0,0,0) within 20 degrees.
# Maximum translation amplitude is set to 0.2A to all defined groups.
for g in ENGINE.groups:
g.set_move_generator( TranslationTowardsCenterGenerator(amplitude=0.2, center={"fixed":(0,0,0)}, axis=0, angle=25) )
direction¶Direction value.
center¶Center value.
angle¶Angle value in rad.
set_direction(direction)¶Set the generated translation vectors direction.
| Parameters: |
|
|---|
set_angle(angle)¶Sets the tolerance maximum angle.
| Parameters: |
|
|---|
set_center(center)¶Set center value.
| Parameters: |
|
|---|
transform_coordinates(coordinates, argument=None)¶Translate coordinates.
| Parameters: |
|
|---|---|
| Returns: |
|
fullrmc.Generators.Translations.generate_random_float()¶random() -> x in the interval [0, 1).
Agitations contains all MoveGenerator classes that agitate and shake structures such as distances, angles, etc.
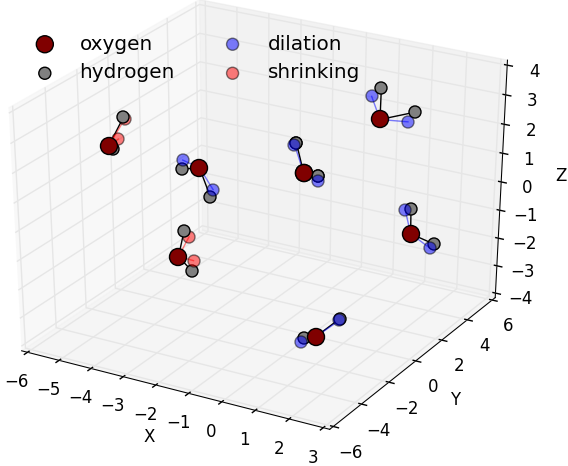
Random H-O-H angle agitation generated on water
molecules. At room temperature, water molecule
angle formed between the two vectors formed between
consecutively the Oxygen atom and the two hydrogen
atoms is about 105 deg. Shrank H-O-H angles are
represented by the red hydrogen while expanded
angles are represented in blue.
( |
fullrmc.Generators.Agitations.DistanceAgitationGenerator(group=None, amplitude=0.2, symmetric=True, shrink=None, agitate=(True, True))¶Bases: fullrmc.Core.MoveGenerator.MoveGenerator
Generates random agitation moves upon a distance separating two atoms by translating both atoms away from each other or closer to each other along the direction line between them. This is mainly used to shake two atoms bond distance by increasing and decreasing the bond length. Only groups of length 2 are accepted.
| Parameters: |
|
|---|
# import fullrmc modules
from fullrmc.Engine import Engine
from fullrmc.Generators.Agitations import DistanceAgitationGenerator
# create engine
ENGINE = Engine(path='my_engine.rmc')
# set pdb file
ENGINE.set_pdb('system.pdb')
# Add constraints ...
# Re-define groups if needed ...
# Re-define groups selector if needed ...
# set moves generators to random agitations of distance seperating two atoms.
# Maximum agitation amplitude is set to to 0.5A
for g in ENGINE.groups:
if len(g)==2:
g.set_move_generator( DistanceAgitationGenerator(amplitude=0.5) )
amplitude¶Maximum agitation amplitude.
shrink¶Shrink flag value.
symmetric¶Symmetric flag value.
agitate¶Agitate tuple flags value.
check_group(group)¶Check the generator’s group.
| Parameters: |
|
|---|
set_amplitude(amplitude)¶Sets maximum translation vector allowed amplitude.
| Parameters: |
|
|---|
set_symmetric(symmetric)¶Set symmetric flag value.
| Parameters: |
|
|---|
set_shrink(shrink)¶Set shrink flag value.
| Parameters: |
|
|---|
set_agitate(agitate)¶Set agitate tuple value.
| Parameters: |
|
|---|
transform_coordinates(coordinates, argument=None)¶Translate coordinates.
| Parameters: |
|
|---|---|
| Returns: |
|
fullrmc.Generators.Agitations.AngleAgitationGenerator(group=None, amplitude=2, symmetric=True, shrink=None, agitate=(True, True))¶Bases: fullrmc.Core.MoveGenerator.MoveGenerator
Generate random agitation moves upon an angle defined between two vectors left-central and right-central where (central, left, right) are three atoms. Move will be performed on left and/or right atom while central atom will always remain fixed. Distances between left/right and central atoms will remain unchanged. This is mainly used to shake bonded atoms angles by increasing and decreasing the bond length. Only groups of length 3 are accepted.
| Parameters: |
|
|---|
# import fullrmc modules
from fullrmc.Engine import Engine
from fullrmc.Generators.Agitations import AngleAgitationGenerator
# create engine
ENGINE = Engine(path='my_engine.rmc')
# set pdb file
ENGINE.set_pdb('system.pdb')
# Add constraints ...
# Re-define groups if needed ...
# Re-define groups selector if needed ...
# set moves generators to random agitations of the angle formed between
# one central atom and other two. Maximum agitation amplitude is set to to 10.
for g in ENGINE.groups:
if len(g)==3:
g.set_move_generator( AngleAgitationGenerator(amplitude=10) )
amplitude¶Maximum agitation angle amplitude in rad.
shrink¶Shrink flag value.
symmetric¶Symmetric flag value.
agitate¶Agitate tuple flags value.
check_group(group)¶Check the generator’s group.
| Parameters: |
|
|---|
set_amplitude(amplitude)¶Set maximum allowed agitation rotation angle amplitude in degrees of left and right atoms separately and transforms it to rad.
| Parameters: |
|
|---|
set_symmetric(symmetric)¶Set symmetric flag value.
| Parameters: |
|
|---|
set_shrink(shrink)¶Set shrink flag value.
| Parameters: |
|
|---|
set_agitate(agitate)¶Set agitate tuple value.
| Parameters: |
|
|---|
transform_coordinates(coordinates, argument=None)¶Translate coordinates.
| Parameters: |
|
|---|---|
| Returns: |
|
fullrmc.Generators.Agitations.generate_random_float()¶random() -> x in the interval [0, 1).
Swaps contains all swap or atoms position exchange MoveGenerator classes.
fullrmc.Generators.Swaps.SwapPositionsGenerator(group=None, swapLength=1, swapList=None)¶Bases: fullrmc.Core.MoveGenerator.SwapGenerator
Generates positional swapping between atoms of the selected group and other atoms randomly selected from swapList.
| Parameters: |
|
|---|
# import fullrmc modules
from fullrmc.Engine import Engine
from fullrmc.Generators.Swaps import SwapPositionsGenerator
# create engine
ENGINE = Engine(path='my_engine.rmc')
# set pdb file
ENGINE.set_pdb('system.pdb')
# Add constraints ...
# Re-define groups if needed ...
# Re-define groups selector if needed ...
##### set swap moves between Lithium and Manganese atoms in Li2MnO3 system #####
# reset engine groups to atoms to insure atomic grouping of all the system's atoms
ENGINE.set_groups_as_atoms()
# get all elements list
elements = ENGINE.allElements
# create list of lithium atoms indexes
liIndexes = [[idx] for idx in xrange(len(elements)) if elements[idx]=='li']
# create list of manganese atoms indexes
mnIndexes = [[idx] for idx in xrange(len(elements)) if elements[idx]=='mn']
# create swap generator to lithium atoms
swapWithLi = SwapPositionsGenerator(swapList=liIndexes)
# create swap generator to manganese atoms
swapWithMn = SwapPositionsGenerator(swapList=mnIndexes)
# set swap generator to groups
for g in ENGINE.groups:
# get group's atom index
idx = g.indexes[0]
# set swap to manganese for lithium atoms
if elements[idx]=='li':
g.set_move_generator(swapWithMn)
# set swap to lithium for manganese atoms
elif elements[idx]=='mn':
g.set_move_generator(swapWithLi)
# the rest are oxygen atoms. Default RandomTranslation generator are kept.
check_group(group)¶Check the generator’s group.
| Parameters: |
|
|---|
set_swap_length(swapLength)¶Set swap length. It will reset swaplist automatically.
| Parameters: |
|
|---|
transform_coordinates(coordinates, argument=None)¶Transform coordinates by swapping. This method is called in every move.
| Parameters: |
|
|---|---|
| Returns: |
|
fullrmc.Generators.Swaps.SwapCentersGenerator(group=None, swapList=None)¶Bases: fullrmc.Core.MoveGenerator.SwapGenerator
Computes geometric center of the selected group, and swaps its atoms by translation to the atoms geometric center of the other atoms which are randomly selected from swapList and vice-versa.
| Parameters: |
|
|---|
# import fullrmc modules
from fullrmc.Engine import Engine
from fullrmc.Generators.Swaps import SwapCentersGenerator
# create engine
ENGINE = Engine(path='my_engine.rmc')
# set pdb file
ENGINE.set_pdb('system.pdb')
# Add constraints ...
# Re-define groups if needed ...
# Re-define groups selector if needed ...
##### set swap moves between first 10 molecular groups of a system #####
# reset engine groups to molecules
ENGINE.set_groups_as_molecules()
# set swap generator to the first 10 groups
GROUPS = [ENGINE.groups[idx] for idx in range(10)]
for gidx, group in enumerate(GROUPS):
swapList = [g.indexes for idx,g in enumerate(GROUPS) if idx!=gidx]
swapGen = SwapCentersGenerator(swapList=swapList)
group.set_move_generator(swapGen)
swapLength¶Swap length. In this Case it is always None as swapLength is not required for this generator.
set_swap_length(swapLength)¶Set swap length. The swap length that defines the length of the group and the length of the every swap sub-list in swapList. It will automatically be set to None as SwapCentersGenerator does not require a fixed length.
| Parameters: |
|
|---|
set_group(group)¶Set the MoveGenerator group.
| Parameters: |
|
|---|
check_group(group)¶Check the generator’s group.
| Parameters: |
|
|---|
transform_coordinates(coordinates, argument=None)¶Transform coordinates by swapping. This method is called in every move.
| Parameters: |
|
|---|---|
| Returns: |
|
fullrmc.Generators.Removes.AtomsRemoveGenerator(group=None, maximumCollected=None, atomsList=None)¶Bases: fullrmc.Core.MoveGenerator.RemoveGenerator
This generator allows removing single atoms at a time from the system. Atoms are randomly picked from atomsList and returned upon calling pick_from_list method.
| Parameters: |
|
|---|
pick_from_list(engine)¶Randomly picks an atom’s index from atomsList. This method checks and verifies maximumCollected prior to picking and index.
| Parameters: |
|
|---|---|
| Returns: |
|